
Laboratory No 201

Lab-Head
Dr. Rolf Daniel
Diploma (Biology), Georg-August University of Göttingen, 1991
Dr. rer. nat. (Microbiology), Georg-August University of Göttingen, 1994
Postdoc (Microbiology), Georg-August University of Göttingen, 1994 -
1995
Postdoc (Cell Biology, Yeast Genetics), University of California,
Berkeley, 1995 -1996
Group leader at the Department of Microbiology and Genetics of the
Georg-August University of Göttingen, since 6/1996
Current
Members of the Lab No. 201 are:
Dr. Rolf Daniel
rdaniel@gwdg.de
Susanne Bowien
susannebowien@hotmail.com
Frank Hoster
fhoster@gwdg.de
Anja Knietsch
akniets@gwdg.de
Baris S. Özyurt
ozyurt23@hotmail.com
Research topics of the lab are:
1) Molecular and biochemical characterization of genes and gene products
involved in the anaerobic conversion of glycerol to 1,3-propanediol by Citrobacter
freundii and Clostridium pasteurianum.
Microorganisms
such as Citrobacter freundii or
Clostridium pasteurianum are able to grow anaerobically on glycerol as sole
carbon and energy source. Glycerol is converted by enteric bacteria to
1,3-propanediol (major product), ethanol, 2,3-butanediol, acetic and lactic
acids. The fermentation pattern of clostridia is different in that they form
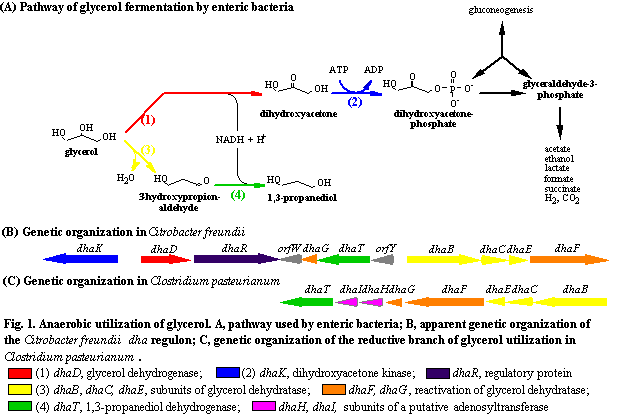
butyric acid and butanol in addition. As outlined in Fig. 1 four
key enzymes are responsible for glycerol breakdown: glycerol dehydrogenase (DhaD),
dihydroxyacetone kinase (DhaK), coenzyme B12-dependent glycerol
dehydratase (DhaB, DhaC, DhaE), and 1,3-propanediol dehydrogenase (DhaT). In our
group, the key enzymes and the corresponding genes of C.
freundii have been identified and characterized. They are encoded by the dha
regulon, the expression of which is induced when dihydroxyacetone or glycerol is
present. In the case of C. pasteurianum,
we have encountered the genetic organization and the gene products of the
reductive branch of glycerol fermentation, which leads to the formation of
1,3-propanediol (Fig. 1).
The
coenzyme B12-dependent glycerol dehydratase is from special interest
because its reaction proceeds via a radical mechanism and its activity is the limiting factor for the biotechnological production of
1,3-propanediol. Our current research is focused on the mechanism of the
reactivation reaction of suicide-inactivated glycerol dehydratase and the
identification of novel dehydratases.
Daniel
R., Gottschalk G. (1992) Growth temperature-dependent activity of glycerol dehydratase in
Escherichia coli expressing the Citrobacter
freundii dha regulon. FEMS
Microbiol. Lett. 100, 281-286.
DANIEL
R., BOENIGK R., GOTTSCHALK G. (1995) Purification of 1,3-propanediol
dehydrogenase from Citrobacter freundii
and cloning, sequencing and overexpression of the corresponding gene in Escherichia coli. J. Bacteriol. 177, 2151-2156.
DANIEL
R., STUERTZ K., GOTTSCHALK G. (1995) Biochemical and molecular characterization
of the oxidative branch of glycerol utilization by Citrobacter
freundii. J. Bacteriol.177, 4392-4401.
SEYFRIED
M., DANIEL R., GOTTSCHALK G. (1996) Cloning, sequencing and overexpression of
the genes encoding coenzyme B12-dependent
glycerol dehydratase of Citrobacter
freundii. J. Bacteriol. 178, 5793-5796.
LUERS
F., SEYFRIED M., DANIEL R., GOTTSCHALK G. (1997) Glycerol conversion to
1,3-propanediol by Clostridium
pasteurianum: cloning and expression of the gene encoding 1,3-propanediol
dehydrogenase. FEMS Microbiol. Lett. 154, 337-345.
MACIS
L., DANIEL R., GOTTSCHALK G. (1998) Properties and sequence of the coenzyme B12-dependent
glycerol dehydratase. FEMS Microbiol. Lett. 164, 21-28.
Daniel
R., BOBIK T. A., Gottschalk G. (1998) Biochemistry of coenzyme B12-dependent
glycerol and diol dehydratases and organization of the encoding genes. FEMS
Microbiol. Rev. 22, 553-566.
Seifert
C, Bowien S, Gottschalk G, Daniel R. (2001)
Identification and expression of the genes and purification and characterization
of the gene products involved in reactivation of coenzyme B12-dependent
glycerol dehydratase of Citrobacter
freundii. European Journal of Biochemistry 268, 2369-2378.
2)
Construction of environmental libraries (metagenomic libraries) and
screening for acquired abilities of the resulting recombinant organisms.
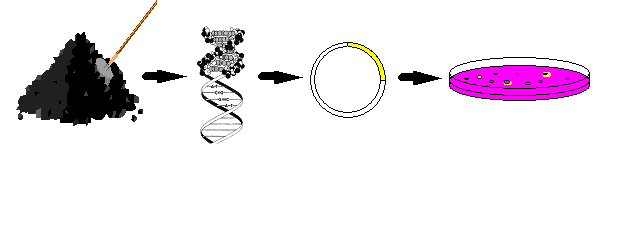
Naturally
occurring assemblages of microorganisms often encompass a bewildering array of
physiological, metabolic, and genetic diversity. It has been estimated that to
date less than 1 % of the worlds organisms can be cultured by using standard
techniques. Therefore, our approach is to use the genetic diversity of the
microorganisms in an environment to encounter new or improved genes and gene
products for biotechnological purposes. Our approach for the identification of
such genes and gene products is outlined in Fig. 2.
HENNE
A., DANIEL R., SCHMITZ R. A., GOTTSCHALK G. (1999) Construction of environmental
DNA libraries in Escherichia coli and
screening for the presence of genes conferring utilization of 4-hydroxybutyrate.
Appl. Environ. Microbiol. 65, 3901-3907.
HENNE
A.,SCHMITZ R. A., BÖMEKE, M., GOTTSCHALK G., DANIEL, R. (2000) Screening of
environmental DNA libraries for the presence of genes lipolytic activity on
Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116.
Majernik
A., Gottschalk G., Daniel R. (2001) Screening of environmental DNA libraries for
the presence of genes conferring Na+ (Li+)/H+
antiporter activity on Escherichia coli:
characterization of the recovered genes and the corresponding gene products. J.
Bacteriol. 183, 6645-6653.
Daniel,
R.
(2002)
Construction of environmental libraries for functional screening of enzyme
activity. In: Directed molecular evolution of proteins. K. Johnson, S. Brakmann
(eds.), pp. 63-78. Wiley-VCH Verlag, Weinheim.
3) Cloning and sequencing of the metagenomes from the Wadden Sea and the
river Leine
Molecular
phylogenetic surveys of different habitats revealed the existence of may new
microbial species and lineages undetected by classical microbiological
approaches. In order to gain insights into genomes of uncharacterized microbial
species, we have started to conduct metagenomic studies employing marine and
freshwater sediments as starting materials (Fig. 3). We have used sediments of
the Wadden Sea and the river Leine as model environments for our studies.
Methods for the direct isolation of high-molecular weight DNA from both habitats
have been developed and genomic fragments have been cloned into cosmid, plasmid,
and BAC vectors. The resulting complex DNA libraries represent a substantial
fraction of the metagenomes of these habitats. These libraries provide access to
the genomic characterization of uncultivated microorganisms. In addition, the
constructed libraries serve as a rich source for the sequence or activity based
identification of novel genes encoding biotechnologically important enzymes and
pathways for the production of bioactive agents .

4)
Other publications
DANIEL
R., WARNECKE F., POTEKHINA J. S., GOTTSCHALK G. (1999) Identification of the
syntrophic partners in a coculture coupling anaerobic methanol oxidation to
Fe(III) reduction. FEMS Microbiol. Lett. 180, 197-203.
Schmitz
R. A., Daniel R., Deppenmeier U., Gottschalk G.
(2001) The anaerobic way of life. In: The Prokaryotes. third edition, B. Balows,
H. G. Trüper, M. Dworkin, W. Harder, K. H. Schleifer (eds.), Springer Verlag,
New York.
Hoster
F., Daniel R., Gottschalk G. (2001) Isolation of a new Thermoanaerobacterium thermosaccharolyticum strain (FH1) producing a
thermostable dextranase. J. Gen. Appl. Microbiol. 47, 187-192.